Introduction
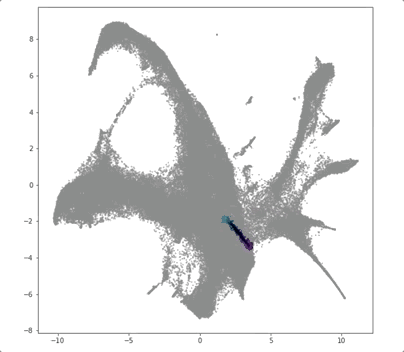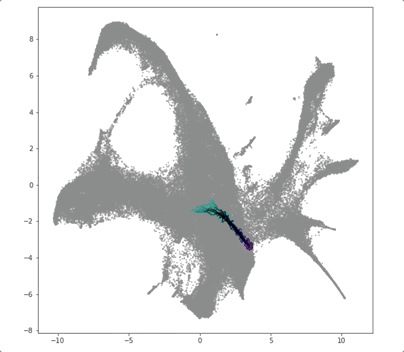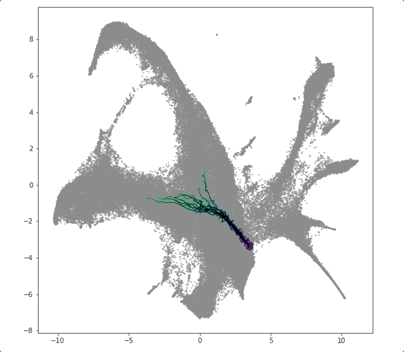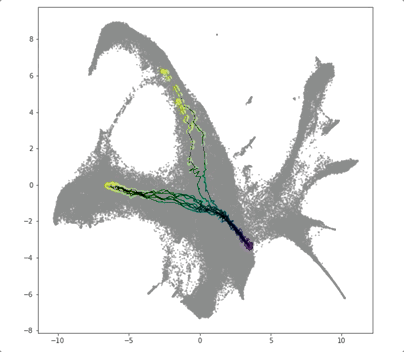
PRESCIENT (Potential eneRgy undeRlying Single-Cell gradIENTs) is a tool for simulating cellular differentiation trajectories with arbitrary cell state intializations. PRESCIENT frames differentiation as a diffusion process given by a stochastic ODE with a drift parameter given by a generative neural network. PRESCIENT models can simulate cellular differentiation trajectories for out-of-sample (i.e. not seen during training) cells, enabling robust fate prediction and perturbational analysis. Here, we package PRESCIENT as a command-line tool and PyPI package.
Installation
We recommend using pip to install PRESCIENT. For a stable version:
pip install prescient
For the latest version:
pip install git+https://github.com/gifford-lab/prescient.git
Source code available at:
github.com/gifford-lab/prescient
Citing
Generative modeling of single-cell time series with PRESCIENT enables prediction of cell trajectories with interventions
by Grace Hui-Ting Yeo, Sachit D. Saksena, and David K. Gifford
Code and notebooks for paper analyses and figures avalable at:
github.com/gifford-lab/prescient-analysis