Interpretability
SIS Interpretability paper is in arXiv
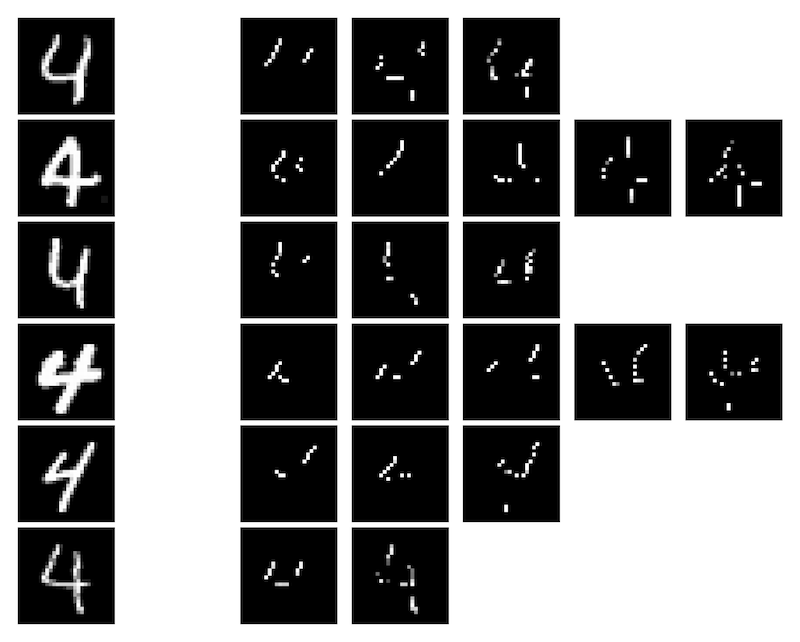
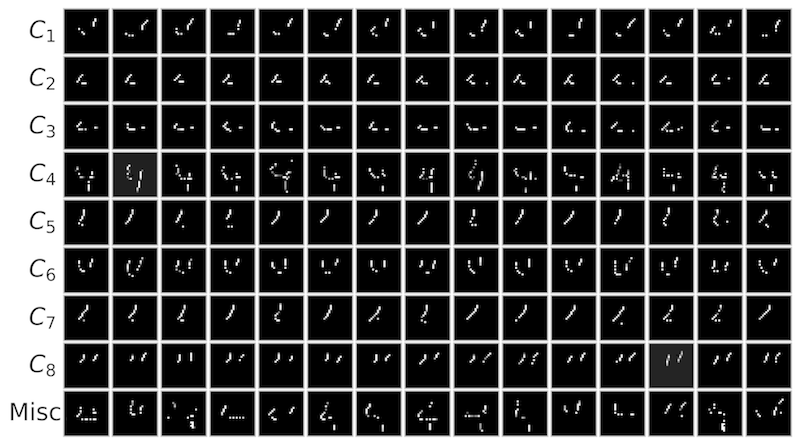
Local explanation frameworks aim to rationalize particular decisions made by a black-box prediction model. Existing techniques are often restricted to a specific type of predictor or based on input saliency, which may be undesirably sensitive to factors unrelated to the model’s decision making process. We instead propose sufficient input subsets that identify minimal subsets of features whose observed values alone suffice for the same decision to be reached, even if all other input feature values are missing. General principles that globally govern a model’s decision-making can also be revealed by searching for clusters of such input patterns across many data points. Our approach is conceptually straightforward, entirely model-agnostic, simply implemented using instance-wise backward selection, and able to produce more concise rationales than existing techniques. We demonstrate the utility of our interpretation method on various neural network models trained on text, image, and genomic data.
Read the paper in arXiv.
SIS applied to two DNA sequences that are positively predicted to bind a specific transcription factor (MAFF) by a CNN:

SIS applied to MNIST digits confidently predicted as class 4 by a CNN: