Reprogramming Recovery
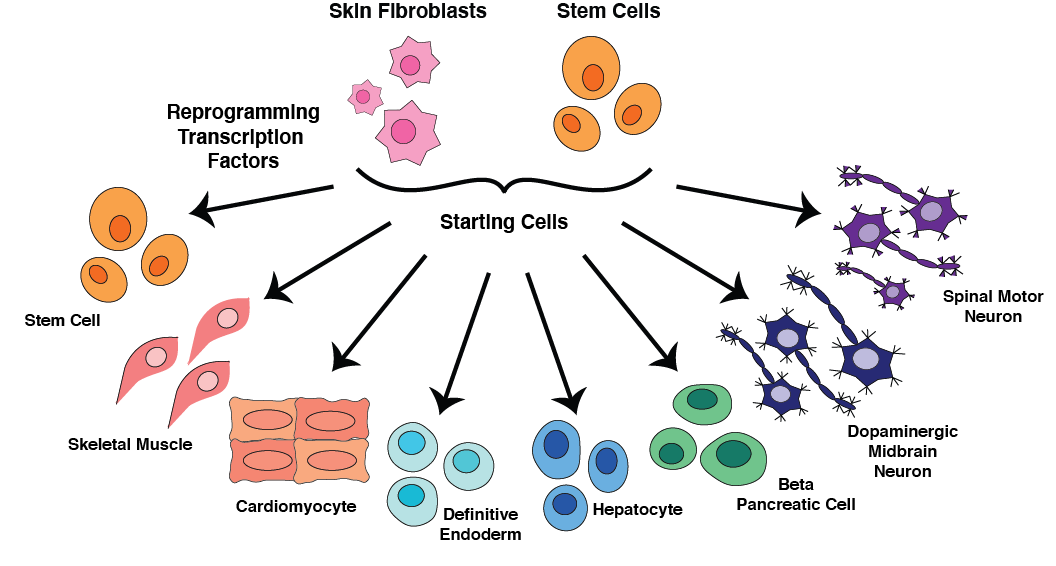
Benchmarking methods for transcription factor prioritization.
View the Project on GitHub gifford-lab/ReprogrammingRecovery
Mouse Multi-tissue Enhancer Sequences
We developed a set of shared multi-tissue mouse enhancers to use as negative control sequences for motif enrichment. We used Mouse ENCODE project candidate enhancer annotations from 18 tissues, where enhancers were defined by both the presence of H3K4me1 and absence of H3K4me3. To be included in our background sequences, the enhancer had to be present in more than 15 out of 18 tissues resulting in a total of 2,609 general enhancers.
Enhancer sequeces are available as a bedfile with mm10 coordinates and as a fasta file.