High-Throughput Interrogation of Gene-Centered Activation Networks
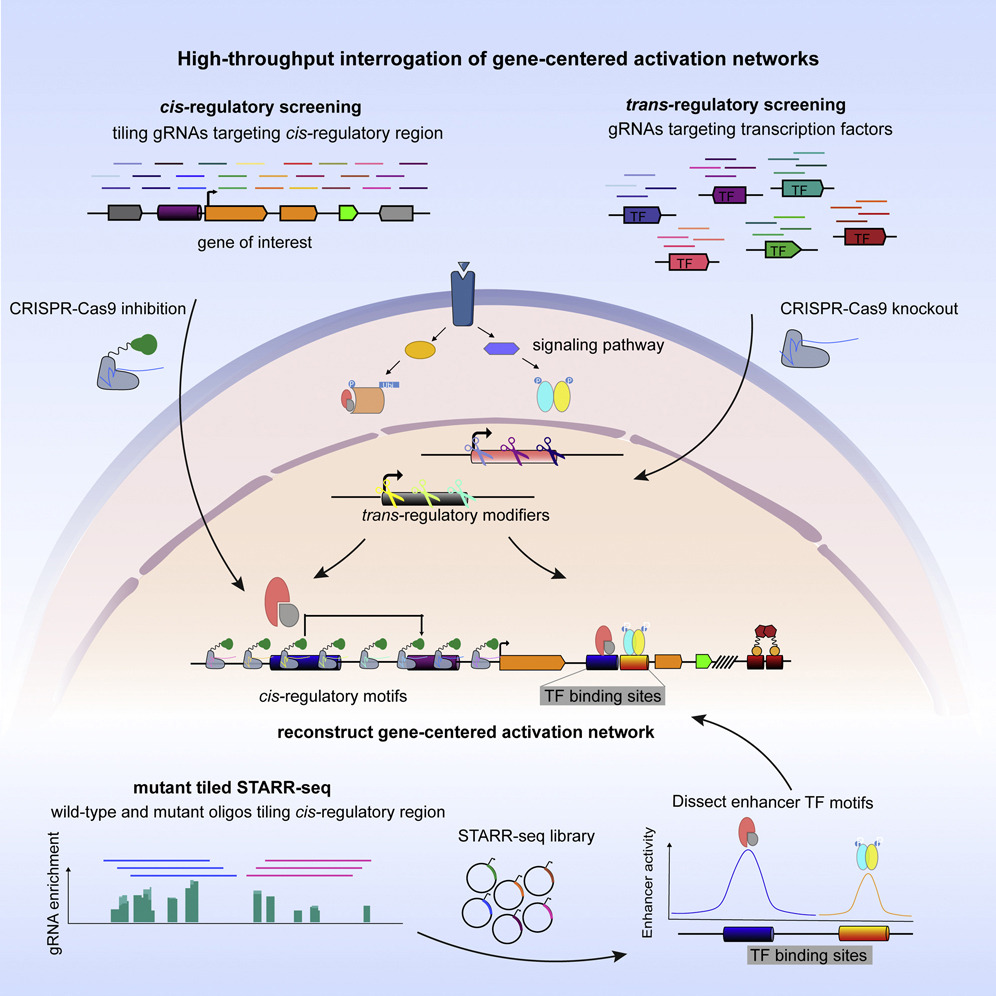
Here we present high-throughput interrogation of gene-centered activation networks (HIGAN), a pipeline that employs a suite of multifaceted genomic approaches to connect upstream signaling inputs, trans-acting TFs, and cis-regulatory elements.
More ›